Demoscript herunterladen (.R)
Demoscript herunterladen (.qmd)
Ordination Hauptkomponentenanalyse (PCA)
Datenbeschreibung
Der Datensatz enthält Daten zum Vorkommen von Fischarten und den zugehörigen Umweltvariablen im Fluss Doubs (Jura). Es gibt 29 Probestellen (sites), an denen jeweils die Abundanzen von 27 Fischarten (auf einer Skalen von 0 bis 5) sowie 10 Umweltvariablen erhoben wurden. In dieser Demo verwenden wir die Umweltdaten:
ele = Elevation (m a.s.l.)
slo = Slope (‰)
dis = Mean annual discharge (m3 s-1)
pH = pH of water
har = Hardness (Ca concentration) (mg L-1)
pho = Phosphate concentration (mg L-1)
nit = Nitrate concentration (mg L-1)
amm = Ammonium concentration (mg L-1)
oxy = Dissolved oxygen (mg L-1)
bod = Biological oxygen demand (mg L-1)
library ("pacman" )p_load ("tidyverse" )# Daten importieren <- read_delim ("./datasets/stat/Doubs_env.csv" , delim = ";" ) |> column_to_rownames (var = "Site" )str (env)
'data.frame': 29 obs. of 10 variables:
$ ele: num 934 932 914 854 849 846 841 752 617 483 ...
$ slo: num 48 3 3.7 3.2 2.3 3.2 6.6 1.2 9.9 4.1 ...
$ dis: num 0.84 1 1.8 2.53 2.64 2.86 4 4.8 10 19.9 ...
$ pH : num 7.9 8 8.3 8 8.1 7.9 8.1 8 7.7 8.1 ...
$ har: num 45 40 52 72 84 60 88 90 82 96 ...
$ pho: num 0.01 0.02 0.05 0.1 0.38 0.2 0.07 0.3 0.06 0.3 ...
$ nit: num 0.2 0.2 0.22 0.21 0.52 0.15 0.15 0.82 0.75 1.6 ...
$ amm: num 0 0.1 0.05 0 0.2 0 0 0.12 0.01 0 ...
$ oxy: num 12.2 10.3 10.5 11 8 10.2 11.1 7.2 10 11.5 ...
$ bod: num 2.7 1.9 3.5 1.3 6.2 5.3 2.2 5.2 4.3 2.7 ...
ele slo dis pH
Min. :172.0 Min. : 0.200 Min. : 0.84 Min. :7.700
1st Qu.:246.0 1st Qu.: 0.500 1st Qu.: 4.80 1st Qu.:7.900
Median :375.0 Median : 1.200 Median :23.00 Median :8.000
Mean :470.9 Mean : 3.531 Mean :22.92 Mean :8.048
3rd Qu.:752.0 3rd Qu.: 3.000 3rd Qu.:28.80 3rd Qu.:8.100
Max. :934.0 Max. :48.000 Max. :69.00 Max. :8.600
har pho nit amm
Min. : 40.00 Min. :0.01 Min. :0.150 Min. :0.0000
1st Qu.: 84.00 1st Qu.:0.10 1st Qu.:0.520 1st Qu.:0.0000
Median : 88.00 Median :0.30 Median :1.600 Median :0.1000
Mean : 85.83 Mean :0.57 Mean :1.697 Mean :0.2124
3rd Qu.: 97.00 3rd Qu.:0.58 3rd Qu.:2.500 3rd Qu.:0.2000
Max. :110.00 Max. :4.22 Max. :6.200 Max. :1.8000
oxy bod
Min. : 4.100 Min. : 1.300
1st Qu.: 8.100 1st Qu.: 2.700
Median :10.200 Median : 4.100
Mean : 9.472 Mean : 5.014
3rd Qu.:11.000 3rd Qu.: 5.200
Max. :12.400 Max. :16.700
PCA durchführen
# Berechnen der PCA <- prcomp (env, scale = TRUE )
Wir verwenden hier «scale = TRUE» weil wir Umweltvariablen mit unterschiedliche Masseinheiten verwenden die variierende Skalen aufweisen. Eine unskalierte PCA (default: scale = FALSE ) ist sinnvoll, wenn alle Variablen ähnliche Skalen haben oder die Skalenunterschiede von Interesse sind.
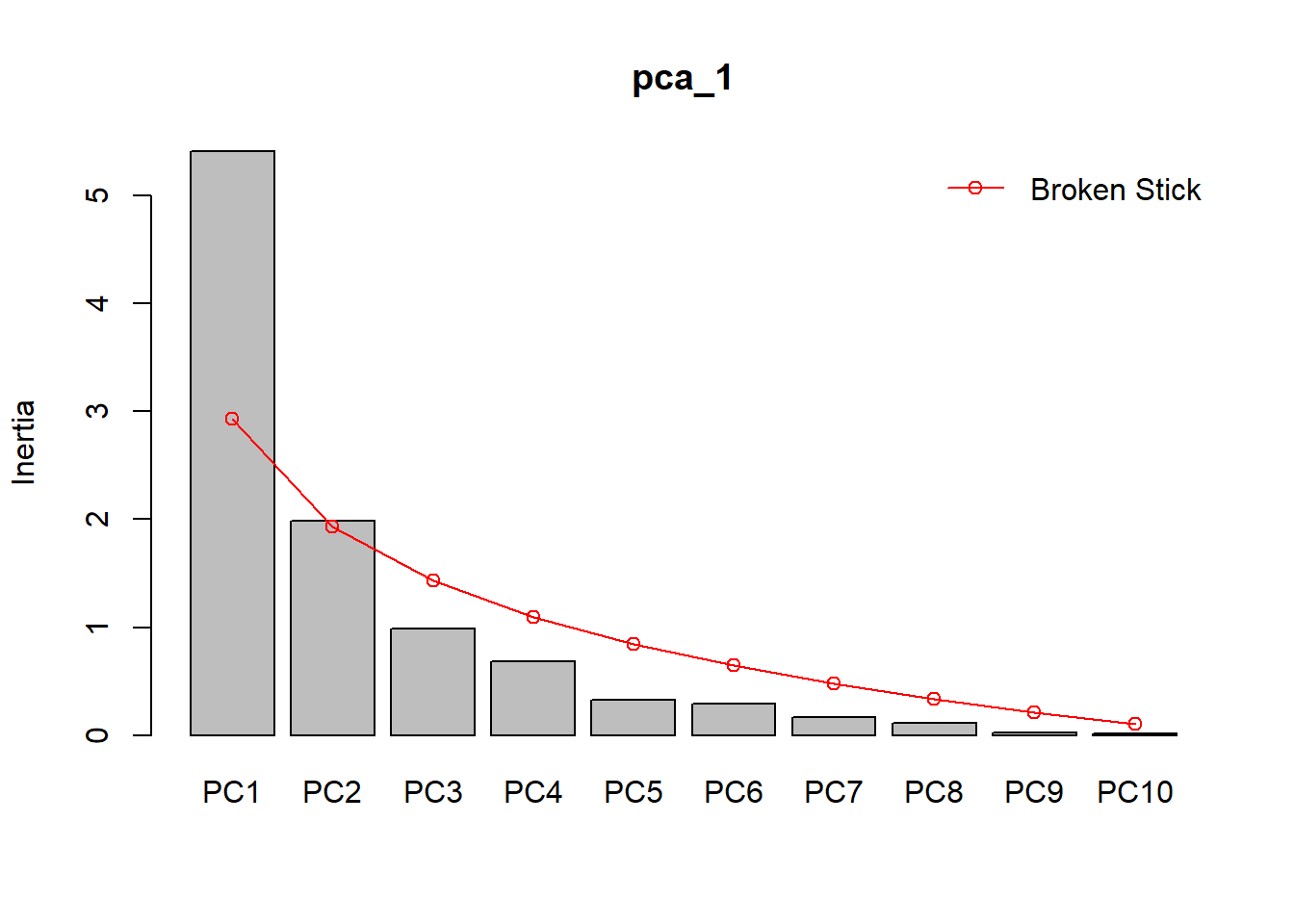
# Erklärte Varianzen der einzelnen Achsen (Principle Components) summary (pca_1)
Importance of components:
PC1 PC2 PC3 PC4 PC5 PC6 PC7
Standard deviation 2.3261 1.4081 0.99377 0.82554 0.57389 0.54229 0.40525
Proportion of Variance 0.5411 0.1983 0.09876 0.06815 0.03293 0.02941 0.01642
Cumulative Proportion 0.5411 0.7393 0.83809 0.90624 0.93917 0.96858 0.98501
PC8 PC9 PC10
Standard deviation 0.33106 0.15186 0.13146
Proportion of Variance 0.01096 0.00231 0.00173
Cumulative Proportion 0.99597 0.99827 1.00000
# Korrelationen der Variablen mit den Ordinationsachsen (Loadings) $ rotation
PC1 PC2 PC3 PC4 PC5 PC6
ele -0.32983498 0.37326508 0.206379458 0.19101532 -0.2108916 -0.13925126
slo -0.18939659 0.36225406 -0.317430615 -0.78012687 -0.2534671 -0.10409281
dis 0.29534136 -0.38102320 -0.289744571 -0.25273405 -0.2684698 0.30100160
pH -0.02798027 -0.30500230 0.833784024 -0.40105127 -0.1951586 0.07666589
har 0.29140671 -0.40748250 -0.128533282 0.05461084 -0.2491737 -0.71799621
pho 0.37752293 0.27210613 0.155636940 -0.10896147 0.1007383 -0.20179074
nit 0.39070251 0.01310715 -0.007977941 -0.20607662 0.5141959 0.23374094
amm 0.36361130 0.32397140 0.159353915 -0.06949906 0.2419581 -0.15450710
oxy -0.35471920 -0.20647070 -0.007373084 -0.25539560 0.5526433 -0.45522763
bod 0.35944351 0.32155396 0.106029560 0.05309465 -0.2837688 -0.17696504
PC7 PC8 PC9 PC10
ele 0.18472542 0.588745894 0.23188511 0.4176369609
slo -0.20463767 0.034575041 -0.01624550 -0.0715819377
dis 0.59712652 0.176111385 0.19635151 0.1759578515
pH -0.07146037 -0.007914091 0.02364683 -0.0214444926
har -0.28531107 0.261864429 0.04422096 0.0001391902
pho 0.34531100 0.146608797 -0.74268910 0.0644251748
nit -0.42339586 0.270495568 0.13496263 0.4636479419
amm 0.21872209 0.152261020 0.50422013 -0.5700251793
oxy 0.36532131 -0.222406219 0.12492546 0.2347031060
bod 0.03358753 -0.620662606 0.25544171 0.4357752560
# # Koordinaten Sites im Ordinationsraum (Scores) $ x
PC1 PC2 PC3 PC4 PC5 PC6
S1 -4.06058121 3.3572582 -1.5353663864 -3.11664281 -0.440940815 0.023962858
S2 -2.85884496 1.6453967 0.6199429169 0.81205916 0.462056595 1.198866058
S3 -2.59500367 0.9087955 1.9223023314 0.08390276 -0.149900842 0.742123168
S4 -2.42274201 0.5759475 0.2416999136 0.73737973 0.197031209 -0.175001441
S5 -0.87891903 1.0132044 0.9242023016 0.92176239 -0.899225211 -0.317177629
S6 -2.07214852 1.4521772 -0.0223558512 1.05958043 -0.018694903 0.245670558
S7 -2.16112216 0.1677976 0.4568713333 0.24335994 -0.322106920 -0.877019196
S8 -0.57361742 0.7209215 0.2639757108 1.22605607 -0.878760741 -0.203164295
S9 -1.35708234 0.9878134 -1.7146160465 0.66588881 -0.007739829 -0.393125521
S10 -0.79292243 -0.8705883 0.0009546047 -0.26509619 0.291231194 -0.650848271
S11 -1.36709366 -0.5303252 -0.8181608908 0.48791374 0.458423383 -0.563740808
S12 -1.22749339 -1.2640408 -0.0337087077 -0.04373209 0.156070410 -0.969587498
S13 -0.78848467 -1.4750236 1.0008336819 -0.52937272 0.125890673 -0.835849370
S14 -1.04150973 -1.8473507 2.4695198950 -1.16235808 -0.117675534 -0.019680069
S15 -0.49079952 -0.5350769 -0.3520651388 0.18345496 0.584620728 -0.025245565
S16 0.24701457 -0.6047699 -0.3687986865 0.13321152 0.579189656 -0.068332189
S17 0.09988521 -0.6508563 -0.3833031874 0.04976758 0.683326378 -0.024162612
S18 0.02438927 -0.7420695 0.1223176607 -0.23010483 0.691984141 0.214472693
S19 0.34858245 -0.6284685 -0.4163658010 -0.13136986 1.081653869 0.335357672
S20 0.25655489 -0.4423705 -0.9691575293 0.38503444 0.366563949 0.530706156
S21 0.14920132 -0.8509911 -0.0692153090 -0.02307061 -0.169105752 0.366684836
S22 4.40893902 1.7764650 1.0312263310 -0.27455931 -0.217479493 -0.607578015
S23 2.99431167 0.7625691 0.0170945415 0.48455983 -0.940755575 0.035682696
S24 7.11385299 3.0888496 0.4707839188 -0.34766485 0.841598956 -0.381766823
S25 2.49964796 0.2144704 -0.8136841889 0.34331209 -0.386714310 0.514435592
S26 1.60318946 -0.6308112 -0.1178042084 -0.23842040 -0.252238973 0.894423496
S27 1.90404098 -1.4844247 0.7045838208 -0.94039312 0.124061006 0.646597704
S28 1.42466912 -1.7701617 -2.2784706379 0.15724184 -0.607940975 0.004076669
S29 1.61408580 -2.3443369 -0.3532363923 -0.67170041 -1.234422275 0.359219148
PC7 PC8 PC9 PC10
S1 -0.16756651 -0.01661397 -0.0005434083 -0.027276628
S2 0.66409559 0.08312574 0.0453575992 -0.114126005
S3 0.36457987 -0.06164832 0.1363572490 0.106940451
S4 0.20481751 0.41948659 -0.1190803292 -0.057120717
S5 -0.33938509 0.29033210 0.1331611406 -0.012913680
S6 0.40988744 -0.32654553 -0.0386629851 0.296369679
S7 -0.11117406 0.48938544 0.0184021086 -0.012862367
S8 -0.67526691 0.44432628 -0.0427742137 -0.107371788
S9 -0.22123560 -0.10940926 -0.0297606988 0.019485234
S10 -0.17677815 0.17051596 -0.0891684003 0.197809147
S11 0.47423174 -0.36666506 0.0305048878 -0.036333811
S12 0.22914572 -0.15537004 0.0333334255 -0.133603754
S13 0.02658378 -0.24507682 0.0275604057 0.219092722
S14 0.17355943 -0.15968840 -0.2347153211 -0.143569558
S15 -0.48268849 -0.02834067 -0.1346994236 -0.055576670
S16 -0.32433001 -0.08783303 0.3085297270 0.140390082
S17 -0.07858369 0.12396263 -0.0949232855 -0.147315115
S18 0.06659440 -0.12827386 -0.2055645165 -0.014781123
S19 -0.24395665 0.13680466 0.2479536264 -0.103423285
S20 -0.31209067 -0.24618792 -0.0271824151 -0.091732053
S21 -0.25594533 -0.46246075 -0.0354205351 -0.180973464
S22 0.24323679 -0.72159949 0.1819244804 0.009669496
S23 -0.39736429 -0.53241824 0.0374850854 -0.145733335
S24 0.43932542 0.60742531 -0.0909990349 -0.048500568
S25 -0.15806719 -0.14778266 -0.4470511938 0.249196626
S26 -0.38708608 -0.06975558 0.1101065003 0.024382817
S27 -0.58223511 0.51151147 0.1409060157 0.199078478
S28 0.88253998 0.27851440 0.1417856127 0.037595473
S29 0.73515615 0.31027906 -0.0028221033 -0.066796286
p_load ("vegan" )# Visualisierung der Anteile erklärter Varianz, im Vergleich zu einem Broken-Stick-Modell screeplot (pca_1, bstick = TRUE )
PCA visualisieren
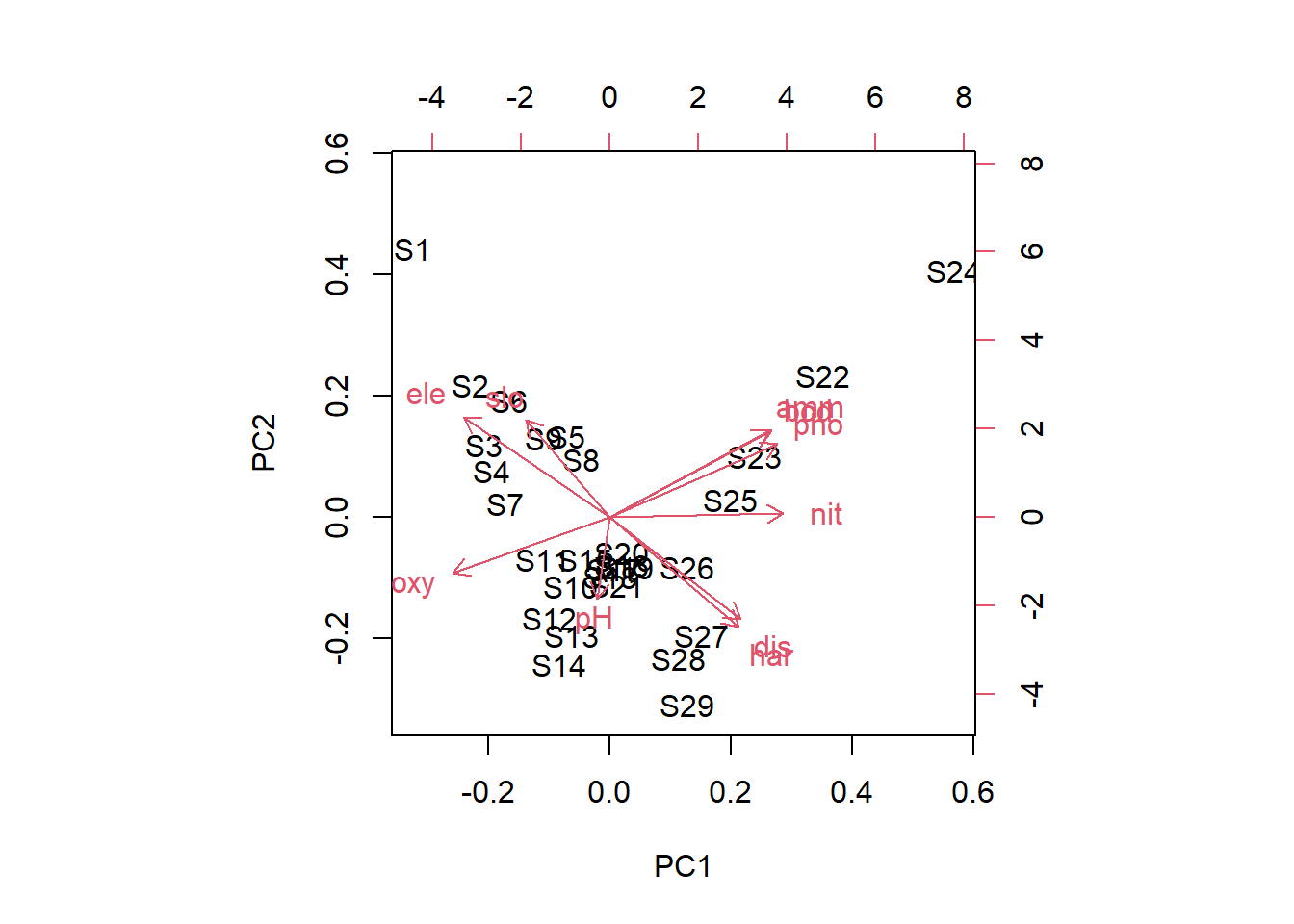
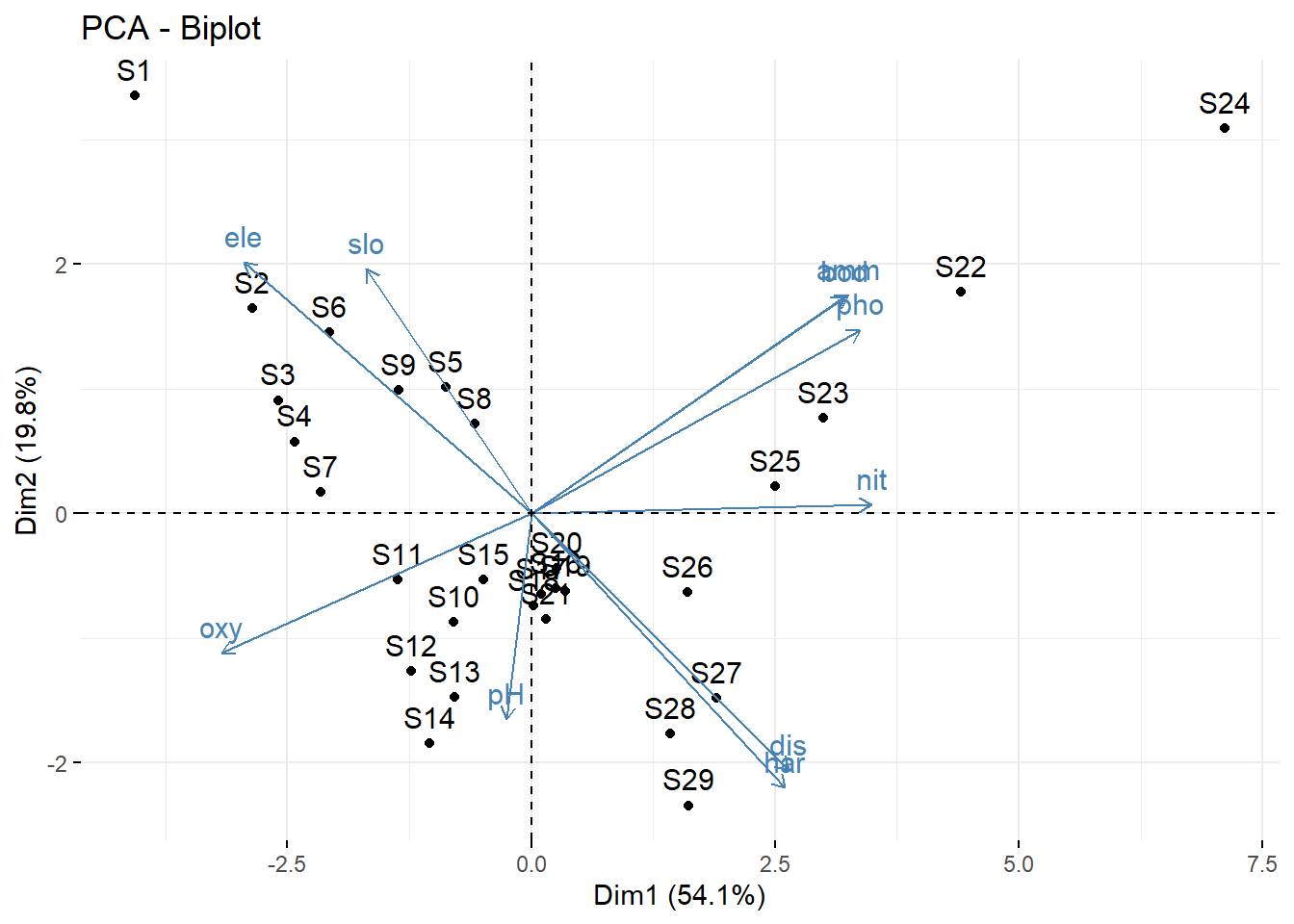
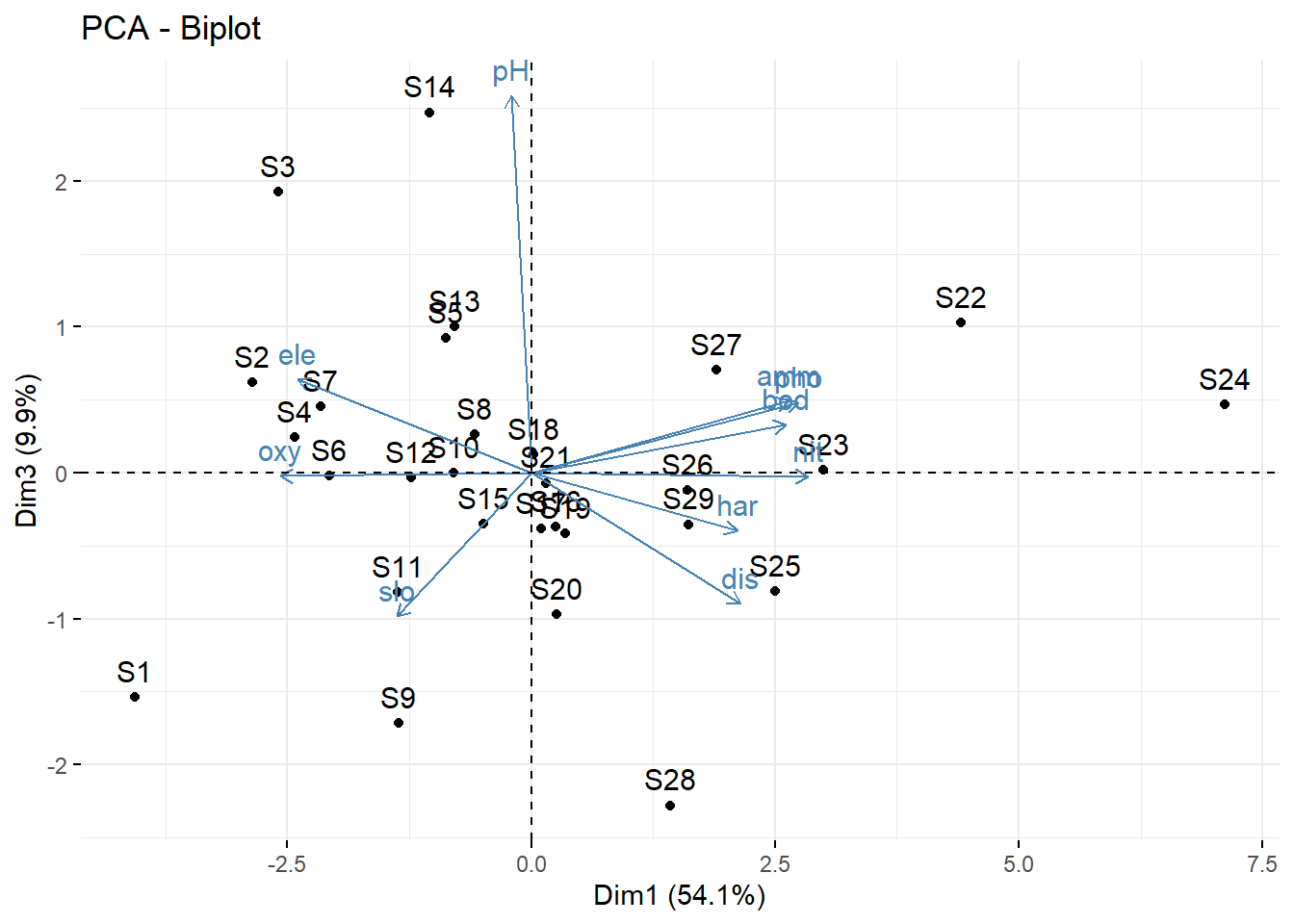
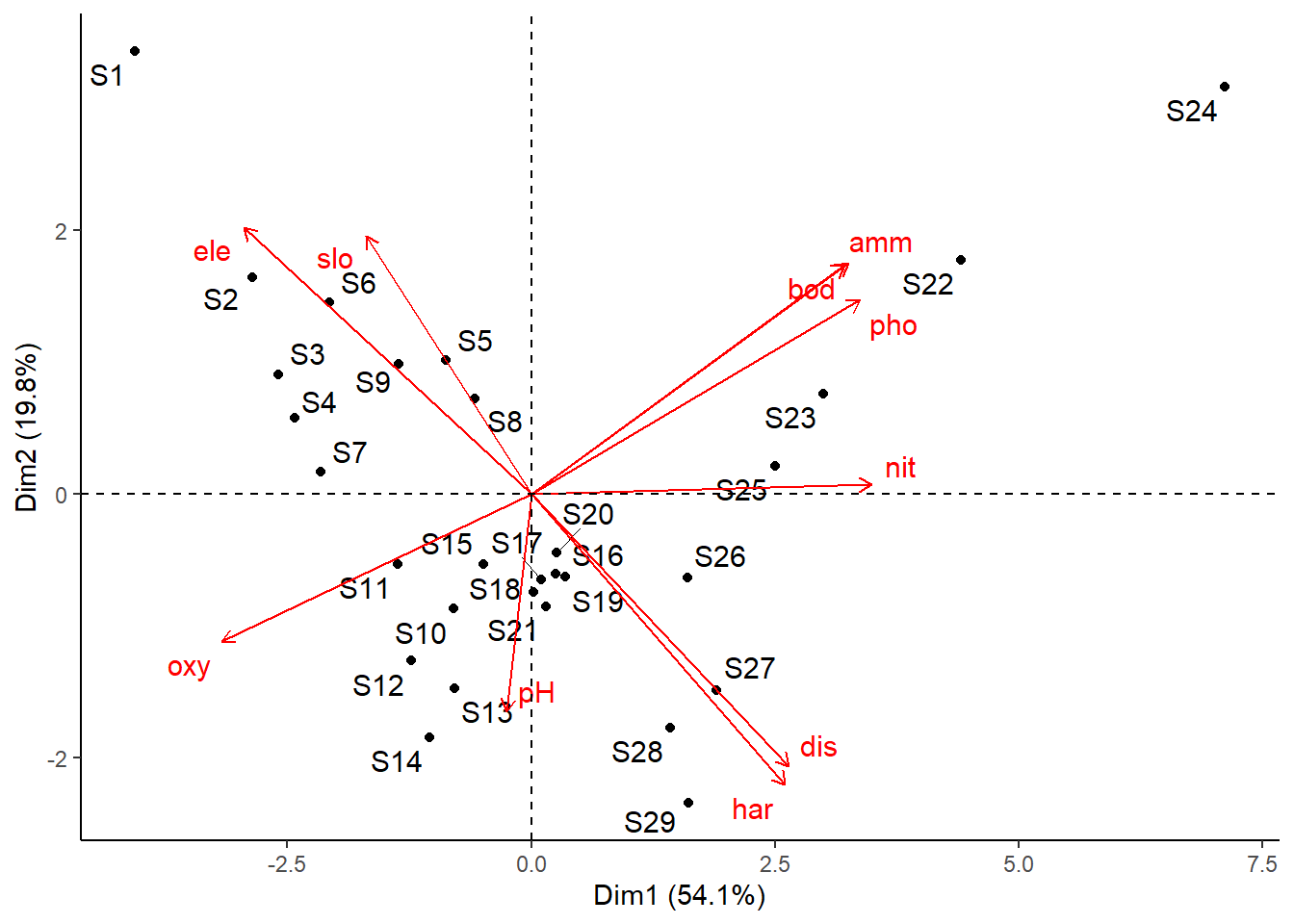
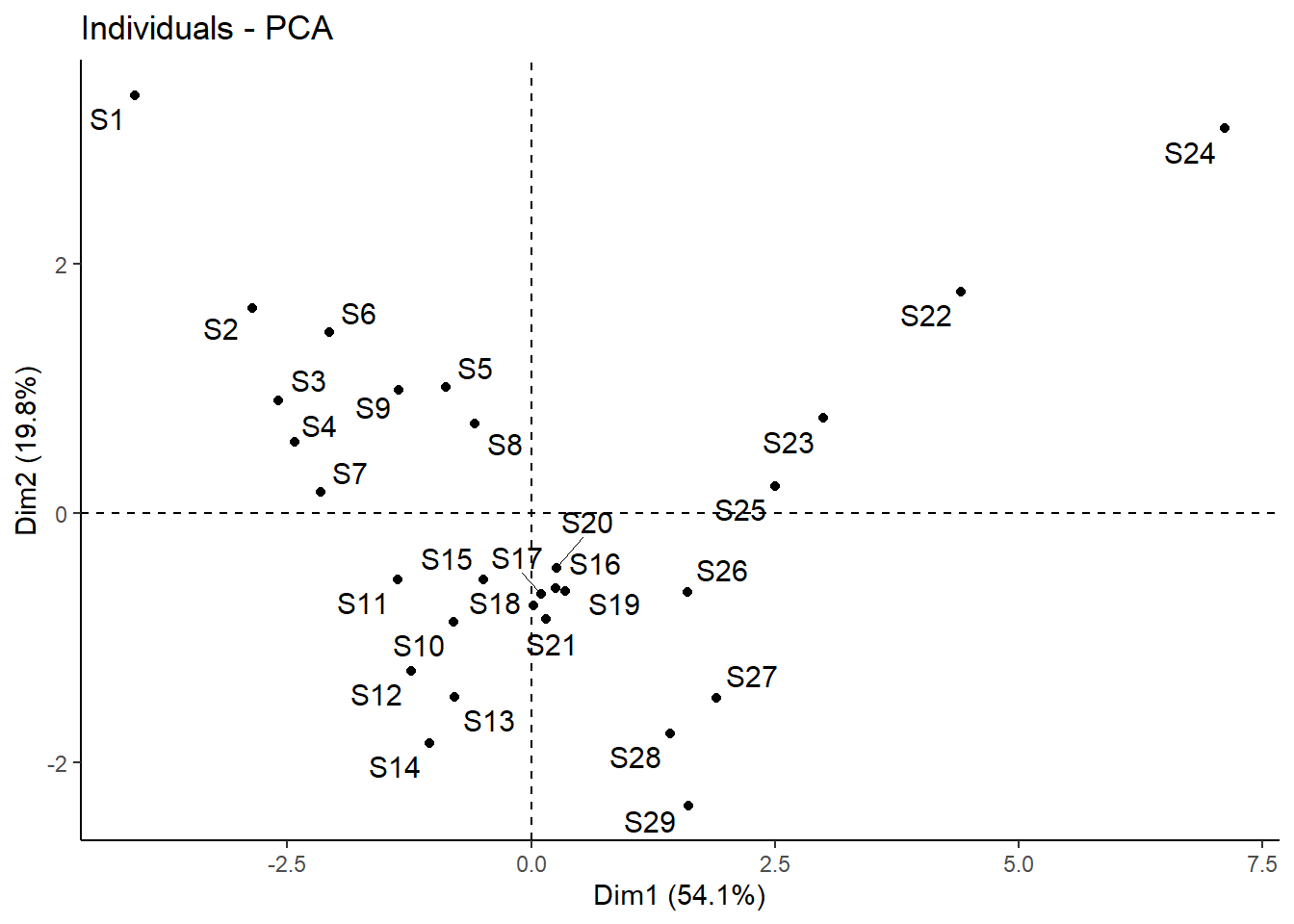
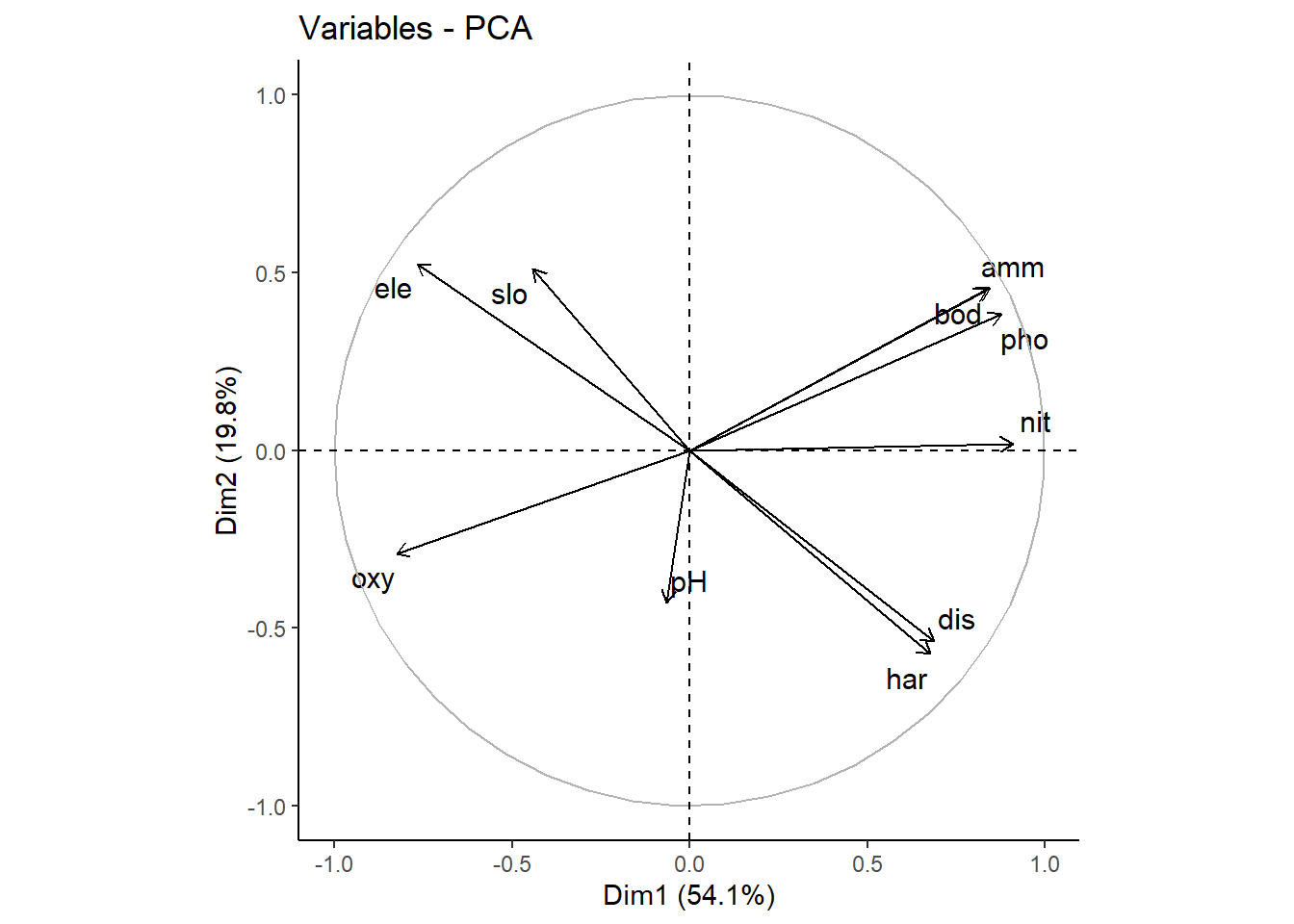
# Mit biplot von base R biplot (pca_1)# Mit package factoextra p_load ("factoextra" )# Biplot fviz_pca_biplot (pca_1)# Biplot der 1 und 3 Achse fviz_pca_biplot (pca_1, axes = c (1 , 3 ) )# Biplot mit angepasssten Grafikparametern # repl = TRUE verhindert die Überlagerung der Textlabels fviz_pca_biplot (pca_1, repel = TRUE , col.var = "red" , col.ind = "black" ) + ggtitle (NULL ) + theme_classic ()# Nur Inidviudals (hier Sites) fviz_pca_ind (pca_1, repel = TRUE ) + theme_classic ()# Nur Variables (hier Umweltparameter) fviz_pca_var (pca_1, repel = TRUE ) + theme_classic ()