# Je 10 Messwerte für Sorte a und b zu einem Data Frame im long-Format verbinden
Messwerte_a <- c(20, 19, 25, 10, 8, 15, 13, 18, 11, 14) # Messwerte von Cultivar a
Messwerte_b <- c(12, 15, 16, 7, 8, 10, 12, 11, 13, 10) # Messwerte von Cultivar b
cultivar <- as.factor( c(rep("a", 10), rep("b", 10))) # Bezeichnug der Cultivare in der Tabelle
blume <- data.frame("cultivar" = cultivar, "size" = c(Messwerte_a, Messwerte_b)) # Data frame erstellen Statistik 1: Demo
Demoscript herunterladen (.qmd)
t-Test
Daten generieren und anschauen
# Boxplots
library(ggplot2)
ggplot(blume, aes(x = cultivar, y = size, fill = cultivar)) +
geom_boxplot() + # Boxplots
geom_dotplot(binaxis = "y", stackdir = "center", alpha = 0.5) # Datenpunkte darstellen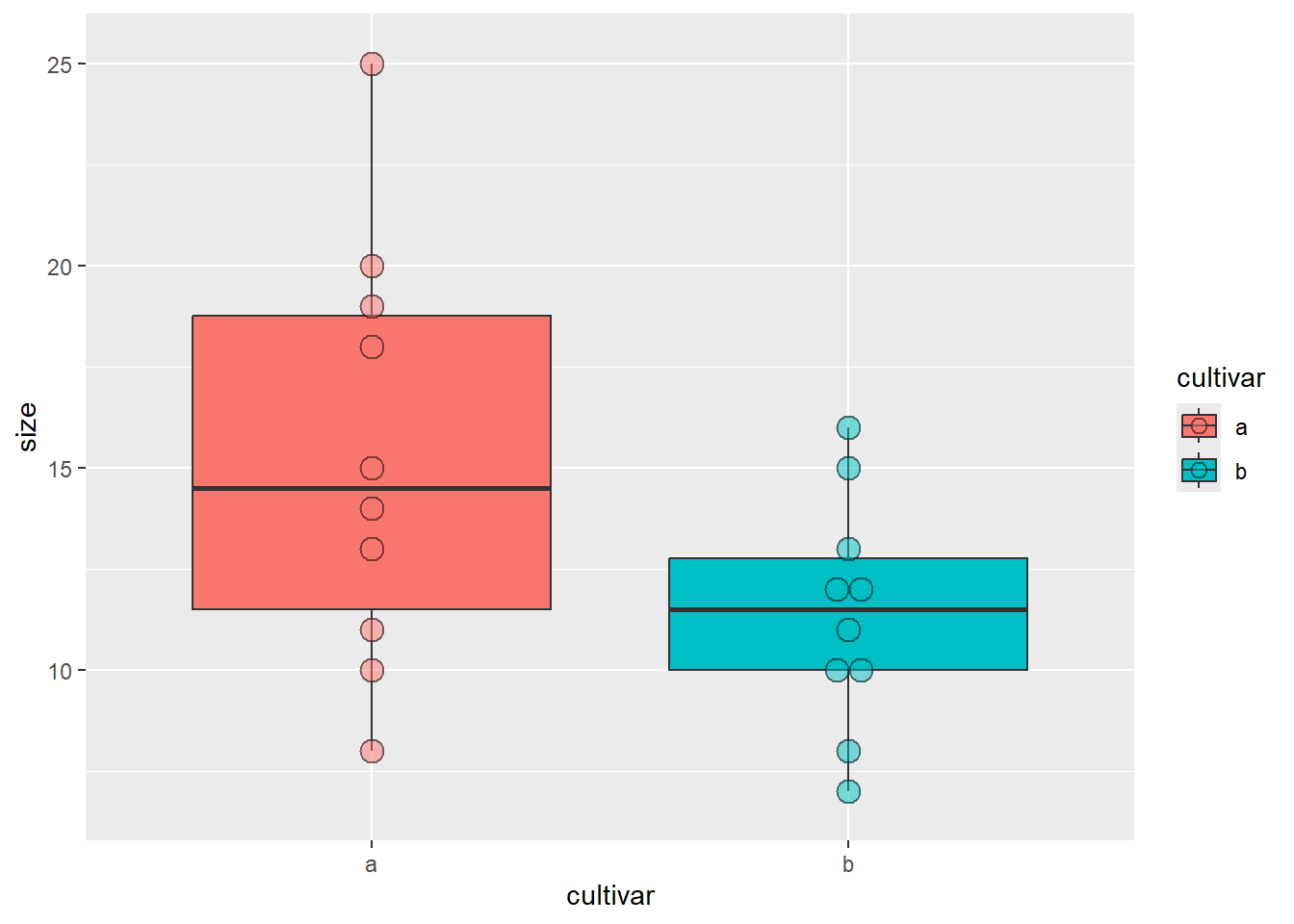
# Histogramme
ggplot(blume, aes(x = size, fill = cultivar)) +
geom_histogram(binwidth = 2) +
facet_wrap(~ cultivar)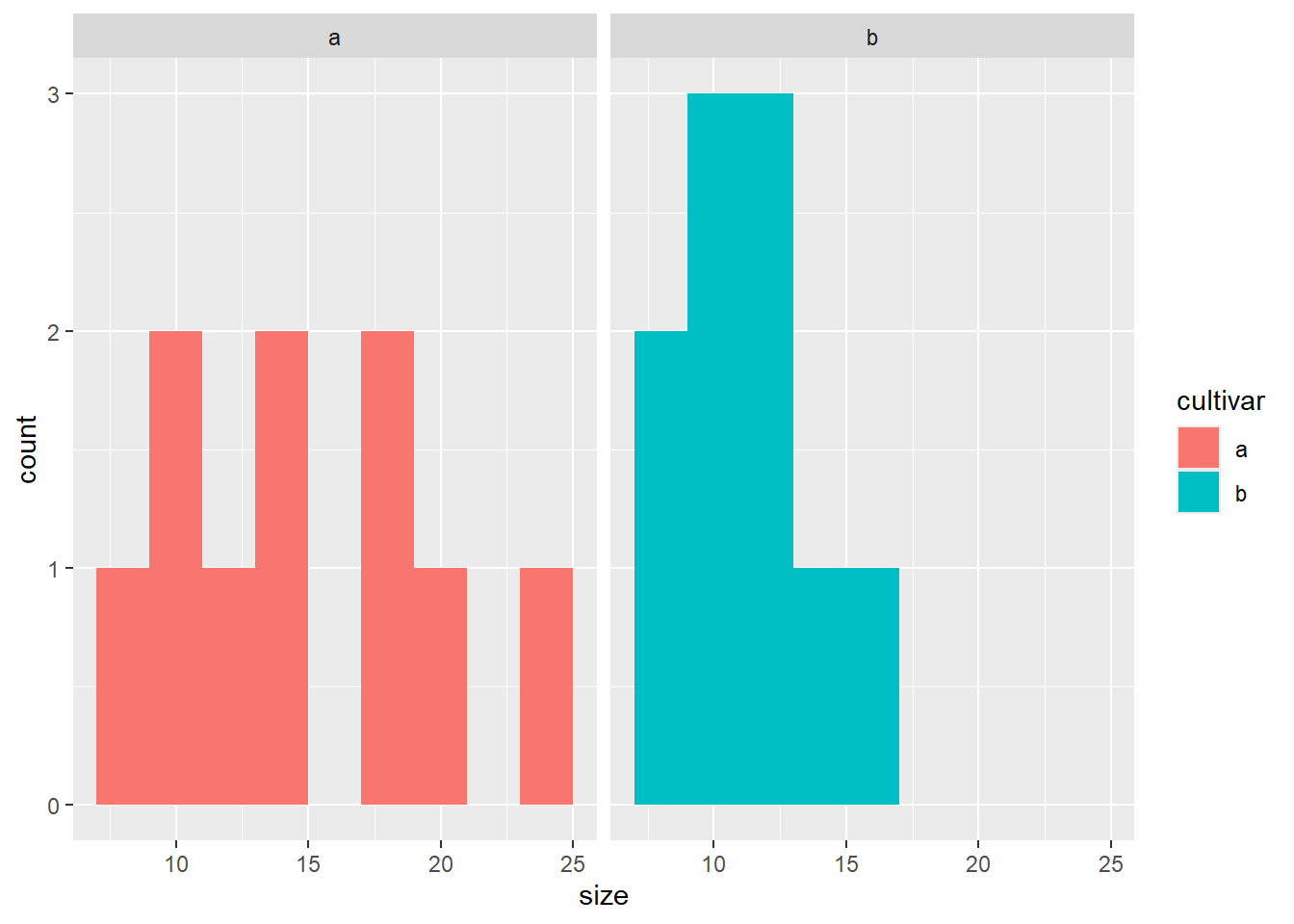
Zweiseitiger t-Test
# Links der Tilde ("~") steht immer die abhängige Variable, rechsts die erklärende(n) Variable(n)
# Alternativ kann man die Werte auch direkt in die t.test()-Funktion eigeben:
# t.test(Messwerte_a, Messwerte_b)
t.test(size ~ cultivar, data = blume) # Zweiseitig "Test auf a ≠ b" (default)
Welch Two Sample t-test
data: size by cultivar
t = 2.0797, df = 13.907, p-value = 0.05654
alternative hypothesis: true difference in means between group a and group b is not equal to 0
95 percent confidence interval:
-0.1245926 7.9245926
sample estimates:
mean in group a mean in group b
15.3 11.4 Einseitiger t-Test
t.test(size ~ cultivar, alternative = "greater", data = blume) # Einseitig "Test auf a > b"
Welch Two Sample t-test
data: size by cultivar
t = 2.0797, df = 13.907, p-value = 0.02827
alternative hypothesis: true difference in means between group a and group b is greater than 0
95 percent confidence interval:
0.5954947 Inf
sample estimates:
mean in group a mean in group b
15.3 11.4 t.test(size ~ cultivar, alternative = "less", data = blume) # Einseitig "Test auf a < b"
Welch Two Sample t-test
data: size by cultivar
t = 2.0797, df = 13.907, p-value = 0.9717
alternative hypothesis: true difference in means between group a and group b is less than 0
95 percent confidence interval:
-Inf 7.204505
sample estimates:
mean in group a mean in group b
15.3 11.4 Klassischer t-Test vs. Welch Test
# Varianzen gleich: klassischer t-Test
t.test(size ~ cultivar, var.equal = TRUE, data = blume)
Two Sample t-test
data: size by cultivar
t = 2.0797, df = 18, p-value = 0.05212
alternative hypothesis: true difference in means between group a and group b is not equal to 0
95 percent confidence interval:
-0.03981237 7.83981237
sample estimates:
mean in group a mean in group b
15.3 11.4 # Varianzen ungleich: Welch's t-Test (siehe Titelzeile des R-Outputs!)
t.test(size ~ cultivar, data = blume) # dasselbe wie var.equal = FALSE
Welch Two Sample t-test
data: size by cultivar
t = 2.0797, df = 13.907, p-value = 0.05654
alternative hypothesis: true difference in means between group a and group b is not equal to 0
95 percent confidence interval:
-0.1245926 7.9245926
sample estimates:
mean in group a mean in group b
15.3 11.4 Gepaarter t-Test
# Gepaarter t-Test: erster Wert von a wird mit erstem Wert von
# b gepaart, zweiter Wert von a mit zweitem von b ect.
t.test(Messwerte_a, Messwerte_b, paired = TRUE) # für gepaarten t-Test funktioniert Notation "size ~ cultivar" nicht
Paired t-test
data: Messwerte_a and Messwerte_b
t = 3.4821, df = 9, p-value = 0.006916
alternative hypothesis: true mean difference is not equal to 0
95 percent confidence interval:
1.366339 6.433661
sample estimates:
mean difference
3.9 Binomialtest
In Klammern übergibt man die Anzahl der Erfolge und die Stichprobengrösse
binom.test(84, 200) # Anzahl Frauen im Nationalrat (≙ 42.0 %; Stand 2019)
Exact binomial test
data: 84 and 200
number of successes = 84, number of trials = 200, p-value = 0.02813
alternative hypothesis: true probability of success is not equal to 0.5
95 percent confidence interval:
0.3507439 0.4916638
sample estimates:
probability of success
0.42 binom.test(116, 200) # Anzahl Männer im Nationalrat (≙ 58.0 %; Stand 2019)
Exact binomial test
data: 116 and 200
number of successes = 116, number of trials = 200, p-value = 0.02813
alternative hypothesis: true probability of success is not equal to 0.5
95 percent confidence interval:
0.5083362 0.6492561
sample estimates:
probability of success
0.58 binom.test(3, 7) # Anzahl Frauen im Bundesrat (≙ 42.9 %; Stand 2019)
Exact binomial test
data: 3 and 7
number of successes = 3, number of trials = 7, p-value = 1
alternative hypothesis: true probability of success is not equal to 0.5
95 percent confidence interval:
0.09898828 0.81594843
sample estimates:
probability of success
0.4285714 Chi-Quadrat-Test & Fishers Test
Ermitteln des kritischen Wertes
Direkter Test in R (dazu Werte als Matrix nötig)
# Matrix mit Haarfarbe&Augenfarbe-Kombinationen erstellen
# 38 blond&blau, 14 dunkel&blau, 11 blond&braun, 51 dunkel&braun
count <- matrix(c(38, 14, 11, 51), nrow = 2)
count # Check [,1] [,2]
[1,] 38 11
[2,] 14 51rownames(count) <- c("blond", "dunkel") # Benennen für Übersicht
colnames(count) <- c("blau", "braun") # Benennen für Übersicht
count # Check blau braun
blond 38 11
dunkel 14 51# Tests durchführen
chisq.test(count)
Pearson's Chi-squared test with Yates' continuity correction
data: count
X-squared = 33.112, df = 1, p-value = 8.7e-09fisher.test(count)
Fisher's Exact Test for Count Data
data: count
p-value = 2.099e-09
alternative hypothesis: true odds ratio is not equal to 1
95 percent confidence interval:
4.746351 34.118920
sample estimates:
odds ratio
12.22697 